SGD variants
Considering a cost functional with the same characteristics as the one used for the SGD case, then
When solving the SGD simulation, the following classes / routines were considered
honsgdSGD
def hosgdSGD(OptProb, nEpoch, blkSize, alpha0, hyperFun): W = OptProb.randSol() stats = np.zeros( [nEpoch, OptProb.Nstats] ) nBlk = np.floor_divide(OptProb.X.shape[1],blkSize) if np.remainder(OptProb.X.shape[1],blkSize) > 0: nBlk += 1 k = 0 for e in range(nEpoch): # Generate permutation blkInd = 0 perm = np.random.permutation(OptProb.X.shape[1]) for b in range(nBlk): # general iteration counter k += 1 # select indexes if b <= nBlk-2: n = perm[blkInd:blkInd+blkSize] else: n = perm[blkInd:OptProb.X.shape[1]] blkInd += blkSize # Compute the gradient g = OptProb.gradFun(W, n) alpha = hyperFun.LR(W, alpha0, e, None) W = W - alpha*g # _END_ for(b) stats[e,:] = OptProb.computeStats(W, alpha, e, g) OptProb.printStats(e, nEpoch, stats) return W, stats
Using the Generalized SGD as a reference (in particular the “\(\mathbf{\mbox{for }} b\)” loop), and assuming \(\beta\) is a given value, then
Using the Generalized SGD as a reference (in particular the “\(\mathbf{\mbox{for }} b\)” loop), and assuming \(\gamma_2\) and \(\epsilon\) are given values, then
Using the Generalized SGD as a reference (in particular the “\(\mathbf{\mbox{for }} b\)” loop), and assuming \(\gamma_1\), \(\gamma_2\) and \(\epsilon\) are given values, then
Proposed solution
Among other pausible alternatives, the considered SGD variants may be written as
Highlighted lines point out the differences between SGD and Momentum [Pol64].
def hosgdMTM(OptProb, nEpoch, blkSize, alpha0, hyperFun, momentum=0.9):
W = OptProb.randSol()
z = 0*W
stats = np.zeros( [nEpoch, OptProb.Nstats] )
nBlk = np.floor_divide(OptProb.X.shape[1],blkSize)
if np.remainder(OptProb.X.shape[1],blkSize) > 0:
nBlk += 1
k = 0
for e in range(nEpoch):
# Generate permutation
blkInd = 0
perm = np.random.permutation(OptProb.X.shape[1])
for b in range(nBlk):
# general iteration counter
k += 1
# select indexes
if b <= nBlk-2:
n = perm[blkInd:blkInd+blkSize]
else:
n = perm[blkInd:OptProb.X.shape[1]]
blkInd += blkSize
# Compute the gradient
g = OptProb.gradFun(W, n)
alpha = hyperFun.LR(W, alpha0, e, None)
z = momentum*z - alpha*g
W = W + z
# _END_ for(b)
stats[e,:] = OptProb.computeStats(W, alpha, e, g)
OptProb.printStats(e, nEpoch, stats)
return W, stats
Highlighted lines point out the differences between SGD and RMSProp [HSS12].
def hosgdRMSProp(OptProb, nEpoch, blkSize, alpha0, hyperFun, gamma2=0.9, epsilon=1e-7):
W = OptProb.randSol()
v = 0*W
stats = np.zeros( [nEpoch, OptProb.Nstats] )
nBlk = np.floor_divide(OptProb.X.shape[1],blkSize)
if np.remainder(OptProb.X.shape[1],blkSize) > 0:
nBlk += 1
k = 0
for e in range(nEpoch):
# Generate permutation
blkInd = 0
perm = np.random.permutation(OptProb.X.shape[1])
for b in range(nBlk):
# general iteration counter
k += 1
# select indexes
if b <= nBlk-2:
n = perm[blkInd:blkInd+blkSize]
else:
n = perm[blkInd:OptProb.X.shape[1]]
blkInd += blkSize
# Compute the gradient
g = OptProb.gradFun(W, n)
alpha = hyperFun.LR(W, alpha0, e, None)
v = gamma2*v +(1-gamma2)*g*g
W = W - ( alpha/np.sqrt(epsilon + v) )*g
# _END_ for(b)
stats[e,:] = OptProb.computeStats(W, alpha, e, g)
OptProb.printStats(e, nEpoch, stats)
return W, stats
Highlighted lines point out the differences between SGD and ADAM [KB15].
def hosgdADAM(OptProb, nEpoch, blkSize, alpha0, hyperFun, gamma1=0.9, gamma2=0.999, epsilon=1e-7):
W = OptProb.randSol()
v = 0*W
z = 0*W
stats = np.zeros( [nEpoch, OptProb.Nstats] )
nBlk = np.floor_divide(OptProb.X.shape[1],blkSize)
if np.remainder(OptProb.X.shape[1],blkSize) > 0:
nBlk += 1
k = 0
for e in range(nEpoch):
# Generate permutation
blkInd = 0
perm = np.random.permutation(OptProb.X.shape[1])
for b in range(nBlk):
# general iteration counter
k += 1
# select indexes
if b <= nBlk-2:
n = perm[blkInd:blkInd+blkSize]
else:
n = perm[blkInd:OptProb.X.shape[1]]
blkInd += blkSize
# Compute the gradient
g = OptProb.gradFun(W, n)
alpha = hyperFun.LR(W, alpha0, e, None)
v = gamma2*v + (1-gamma2)*g*g
z = gamma1*z + (1-gamma1)*g
W = W - alpha*(1./(np.sqrt(v) + epsilon) )*( np.sqrt(1-np.power(gamma2,float(k)))/(1-np.power(gamma1,float(k))) )*g
# _END_ for(b)
stats[e,:] = OptProb.computeStats(W, alpha, e, g)
OptProb.printStats(e, nEpoch, stats)
return W, stats
Full solution
HoSGDlabSGDVariants.py
import numpy as np
import scipy as sc
from scipy import signal
from scipy.special import softmax,logsumexp
import matplotlib.pylab as PLT
from matplotlib.ticker import MaxNLocator
import HoSGDdefs as hosgdDef
from HoSGDdefs import hosgdOptProb
import os
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '2' # Disbales TF's warnings (they don' shown in generated HTML)
# Note: TF is used to have a simple access to MNIST
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.datasets import mnist
from tensorflow.keras.datasets import cifar10
from HoSGDlabSGD import hosgdFunSoftMax, hosgdHyperFunSGD, hosgdSGD, stdPreproc
def hosgdMTM(OptProb, nEpoch, blkSize, alpha0, hyperFun, momentum=0.9):
W = OptProb.randSol()
z = 0*W
stats = np.zeros( [nEpoch, OptProb.Nstats] )
nBlk = np.floor_divide(OptProb.X.shape[1],blkSize)
if np.remainder(OptProb.X.shape[1],blkSize) > 0:
nBlk += 1
k = 0
for e in range(nEpoch):
# Generate permutation
blkInd = 0
perm = np.random.permutation(OptProb.X.shape[1])
for b in range(nBlk):
# general iteration counter
k += 1
# select indexes
if b <= nBlk-2:
n = perm[blkInd:blkInd+blkSize]
else:
n = perm[blkInd:OptProb.X.shape[1]]
blkInd += blkSize
# Compute the gradient
g = OptProb.gradFun(W, n)
alpha = hyperFun.LR(W, alpha0, e, None)
z = momentum*z - alpha*g
W = W + z
# _END_ for(b)
stats[e,:] = OptProb.computeStats(W, alpha, e, g)
OptProb.printStats(e, nEpoch, stats)
return W, stats
def hosgdRMSProp(OptProb, nEpoch, blkSize, alpha0, hyperFun, gamma2=0.9, epsilon=1e-7):
W = OptProb.randSol()
v = 0*W
stats = np.zeros( [nEpoch, OptProb.Nstats] )
nBlk = np.floor_divide(OptProb.X.shape[1],blkSize)
if np.remainder(OptProb.X.shape[1],blkSize) > 0:
nBlk += 1
k = 0
for e in range(nEpoch):
# Generate permutation
blkInd = 0
perm = np.random.permutation(OptProb.X.shape[1])
for b in range(nBlk):
# general iteration counter
k += 1
# select indexes
if b <= nBlk-2:
n = perm[blkInd:blkInd+blkSize]
else:
n = perm[blkInd:OptProb.X.shape[1]]
blkInd += blkSize
# Compute the gradient
g = OptProb.gradFun(W, n)
alpha = hyperFun.LR(W, alpha0, e, None)
v = gamma2*v +(1-gamma2)*g*g
W = W - ( alpha/np.sqrt(epsilon + v) )*g
# _END_ for(b)
stats[e,:] = OptProb.computeStats(W, alpha, e, g)
OptProb.printStats(e, nEpoch, stats)
return W, stats
def hosgdADAM(OptProb, nEpoch, blkSize, alpha0, hyperFun, gamma1=0.9, gamma2=0.999, epsilon=1e-7):
W = OptProb.randSol()
v = 0*W
z = 0*W
stats = np.zeros( [nEpoch, OptProb.Nstats] )
nBlk = np.floor_divide(OptProb.X.shape[1],blkSize)
if np.remainder(OptProb.X.shape[1],blkSize) > 0:
nBlk += 1
k = 0
for e in range(nEpoch):
# Generate permutation
blkInd = 0
perm = np.random.permutation(OptProb.X.shape[1])
for b in range(nBlk):
# general iteration counter
k += 1
# select indexes
if b <= nBlk-2:
n = perm[blkInd:blkInd+blkSize]
else:
n = perm[blkInd:OptProb.X.shape[1]]
blkInd += blkSize
# Compute the gradient
g = OptProb.gradFun(W, n)
alpha = hyperFun.LR(W, alpha0, e, None)
v = gamma2*v + (1-gamma2)*g*g
z = gamma1*z + (1-gamma1)*g
W = W - alpha*(1./(np.sqrt(v) + epsilon) )*( np.sqrt(1-np.power(gamma2,float(k)))/(1-np.power(gamma1,float(k))) )*g
# _END_ for(b)
stats[e,:] = OptProb.computeStats(W, alpha, e, g)
OptProb.printStats(e, nEpoch, stats)
return W, stats
Routines to execute example and plot results
def exMultiClass(nEpoch, blkSize, alpha, dataset='mnist', lrPolicy=hosgdDef.lrSGD.Cte, lrSDecay=20, lrTau = 0.5, verbose=5):
# Examples
# w, stats = H.exMultiClass(20, 32, 0.05, dataset='mnist')
# w, stats = H.exMultiClass(20, 32, 0.01, dataset='cifar10')
# w, stats = H.exMultiClass(25, 64, 0.02, dataset='cifar10', lrPolicy=hosgdDef.lrSGD.StepDecay, lrSDecay=10, lrTau=0.5)
# --- Load data
# -------------------
flagDataset = False
if dataset.lower() == 'mnist':
(X, Y), (Xtest, Ytest) = mnist.load_data()
flagDataset = True
if dataset.lower() == 'cifar10':
(X, Y), (Xtest, Ytest) = cifar10.load_data()
flagDataset = True
if flagDataset is False:
print('Dataset {} is no valid'.format(dataset))
return
if verbose == 1:
print('Shape (training data):', X.shape)
print('Shape (valid/testing data):', Xtest.shape)
print('\n')
# pre-processing
X = stdPreproc(X)
Xtest = stdPreproc(Xtest)
# Data vectorization
X = np.transpose( X.reshape(X.shape[0], np.array(X.shape[1::]).prod()) )
Xtest = np.transpose( Xtest.reshape(Xtest.shape[0], np.array(Xtest.shape[1::]).prod()) )
Y = Y.ravel()
Ytest = Ytest.ravel()
# --- Optimization model
# ----------------------
OptProb = hosgdFunSoftMax(X, Y, 10, verbose=verbose, Xtest=Xtest, Ytest=Ytest)
# --- HyperPar (this is kept in order to have a similar structure to the SD case (see Lab 1a)
# ------------
hyperP = hosgdHyperFunSGD(lrPolicy=lrPolicy, lrSDecay=lrSDecay, lrTau=lrTau)
W = []
stats = []
nameVar = []
# --- Call SGD
# -----------
sol = hosgdSGD(OptProb, nEpoch, blkSize, alpha, hyperP)
W.append(sol[0])
stats.append(sol[1])
nameVar.append('SGD')
# --- Call MTM
# -----------
sol = hosgdMTM(OptProb, nEpoch, blkSize, alpha, hyperP)
W.append(sol[0])
stats.append(sol[1])
nameVar.append('Momentum')
# --- Call RMSProp
# -----------
sol = hosgdRMSProp(OptProb, nEpoch, blkSize, alpha, hyperP)
W.append(sol[0])
stats.append(sol[1])
nameVar.append('RMSProp')
# --- Call ADAM
# -----------
sol = hosgdADAM(OptProb, nEpoch, blkSize, alpha, hyperP)
W.append(sol[0])
stats.append(sol[1])
nameVar.append('ADAM')
# --- Plot results
# ----------------
plotSGDStatsList(stats, nameVar)
return W, stats
def plotSGDStatsList(stats,nameVar):
# --- Plot results ---
L = len(stats)
ax = PLT.figure().gca()
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
for k in range(L):
PLT.plot(stats[k][:,1], label=r'{0}'.format(nameVar[k]) )
PLT.legend(loc='upper right')
PLT.ylabel('Softmax', fontsize=16)
PLT.xlabel('Epoch', fontsize=16)
PLT.title('Loss function', fontsize=20)
if stats[0][0,4] is not None:
ax = PLT.figure().gca()
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
for k in range(L):
PLT.plot(stats[k][:,4] )
PLT.ylabel('Accuracy', fontsize=16)
PLT.xlabel('Epoch', fontsize=16)
PLT.title('Success rate (test)', fontsize=20)
PLT.show()
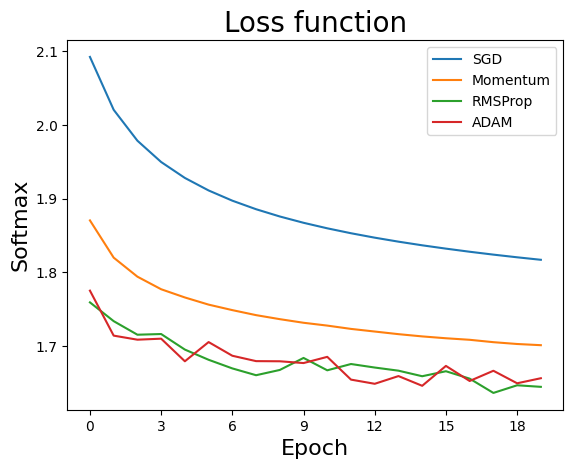
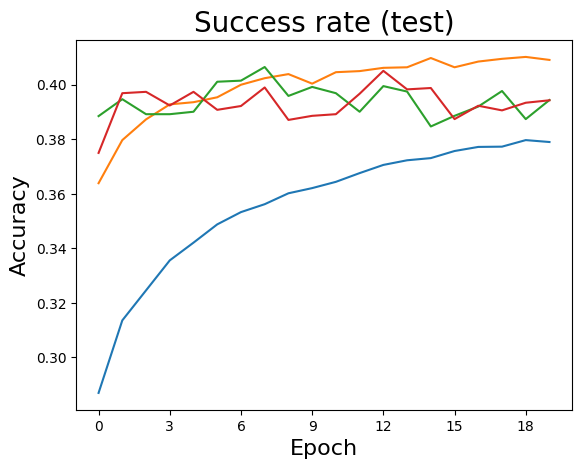
Simple Simulation (MNIST)
import HoSGDlabSGDVariants as SGDvar
# MNIST
# exMultiClass: explicitly executes SGD, SGD+M, RMSProp and ADAM all with the
# same parameters.
sol = SGDvar.exMultiClass(20, 64, 0.001, dataset='mnist',verbose=-1)
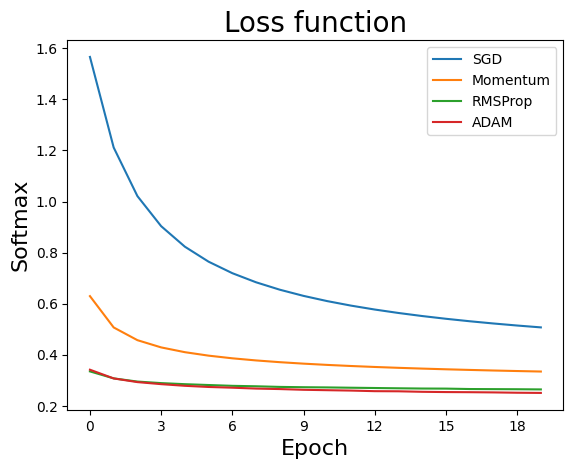
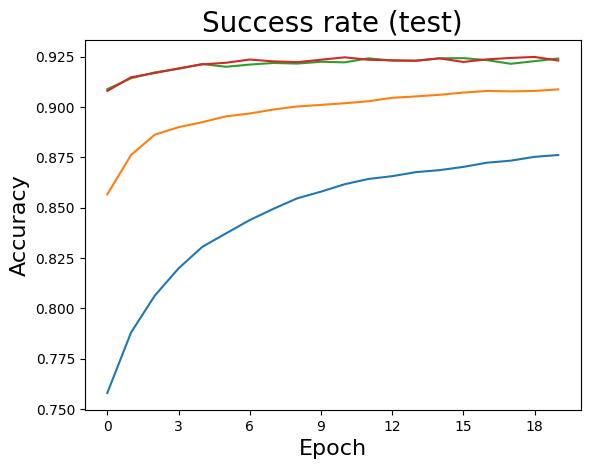
Simple Simulation (CIFAR-10)
import HoSGDlabSGDVariants as SGDvar
# MNIST
# exMultiClass: explicitly executes SGD, SGD+M, RMSProp and ADAM all with the
# same parameters.
sol = SGDvar.exMultiClass(20, 128, 0.001, dataset='cifar10',verbose=-1)